mRNA Signalstructure Detection by RIBL
(GMD)
| Application domain: |
Computational Biology |
| Source: |
Center for Computing Technology (TZI);
University of Bremen |
| Dataset size: |
57 signalstructures (618 ground facts) |
| Data format: |
Prolog |
| Systems Used: |
RIBL |
| Pointers: |
Contact Tamás Horváth
(tamas.horvath@gmd.de) |
The data (GMD)
mRNA is a sequence of nucleotic acids (guanine, adenine, uracil, cytosine).
The secondary structure of an mRNA contains special subsequences called
signalstructures that are responsible for special biological functions
e.g., RNA-protein interactions, poly-A processing, cellular transport etc.
For example the signalstructure Iron Responsive Element (IRE) is responsible
for
- binding of IRE binding protein (aconitase),
- iron depending control of translation and
- cellular switch between iron uptake and iron storage.
Signalstructures have structural motifes that are composed of different building
blocks as e.g., stacking region, hairpin loops, interiror loops etc.
There are similar signalstructures that have the same biological function.
For example 15 different IRE signalstructures have been described by now.
Their similarity roots in a common structure called consensus pattern.
The aims are to find
- new signalstructures of the known classes,
- new signalstructure classes.
RIBL (Emde and Wettschereck, 96) is a relational instance based learning
system developed by GMD. RIBL applies the 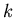 -nearest neighbor method in a
relational representation. As a first step, RIBL collects literals that
describe an object. This is a recursive process in which the depth is
bounded by a user defined parameter.
In the second step, RIBL computes the similarity between objects. Similarity
between objects is computed recursively, similarity between attributes is
defined same as for the propositional case. Finally, the
-nearest neighbor method in a
relational representation. As a first step, RIBL collects literals that
describe an object. This is a recursive process in which the depth is
bounded by a user defined parameter.
In the second step, RIBL computes the similarity between objects. Similarity
between objects is computed recursively, similarity between attributes is
defined same as for the propositional case. Finally, the 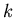 most similar
instances vote on the class of a query. The vote is weighted by similarity.
most similar
instances vote on the class of a query. The vote is weighted by similarity.
We have extended RIBL by a new type for representing sequences. We have
also introduced a similarity measure between sequences. The base of this
similarity measure is the weighted edit distance. The next four target
predicates have been used in this application:
ire/3, tar/3, secis1/3 and
secis2/3.
The fact ire(s_ire1) means, that the class of the
signalstructure s_ire1 is IRE, it has 6 structure
elements and the length of the sequence is 34.
The background predicates hairpin/3, stem/4,
bulge_left/3 and bulge_right/3
represent the structure elements of the signalstructures.
For example the fact hairpin(s_ire1, 1, [c,a,g,t,g,t])
denotes that
the first structure element of the signalstructure
s_ire1 is a hairpin loop with the sequence cagtgt.
Experiments with RIBL (GMD)
The signalstructure database that is currently available for us contains only
the above four signalstructure classes and 57 signalstructures with known
classes. RIBL has 54 signalstructures correctly classified. The 1 and the 2
nearest neighbors have been the best to classify the queries.